Emeriti
Group Glockshuber
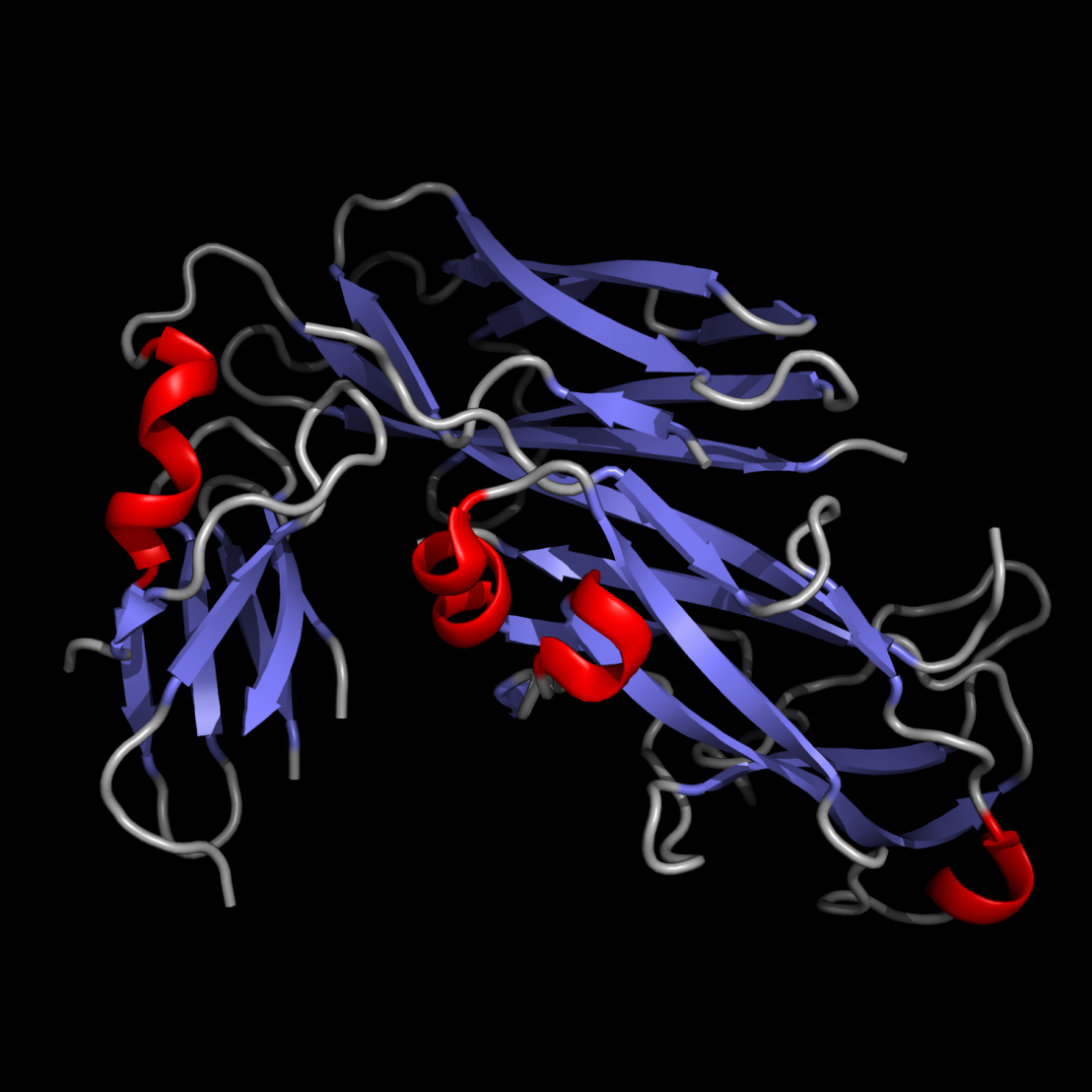
The three-dimensional structure of a protein is exclusively determined by its amino acid sequence and is formed in an autonomous and spontaneous process. Compared to protein folding in vitro, the kinetics and the yields of folding in the living cell are significantly increased by enzymes catalyzing rate-limiting folding steps or preventing nonproductive folding reactions. We are trying to answer general questions on protein folding with particular focus on the mechanism and structural biology of catalyzed disulfide bond formation and the assembly of adhesive type 1 pili in bacteria. Read more about our research.
Group Richmond
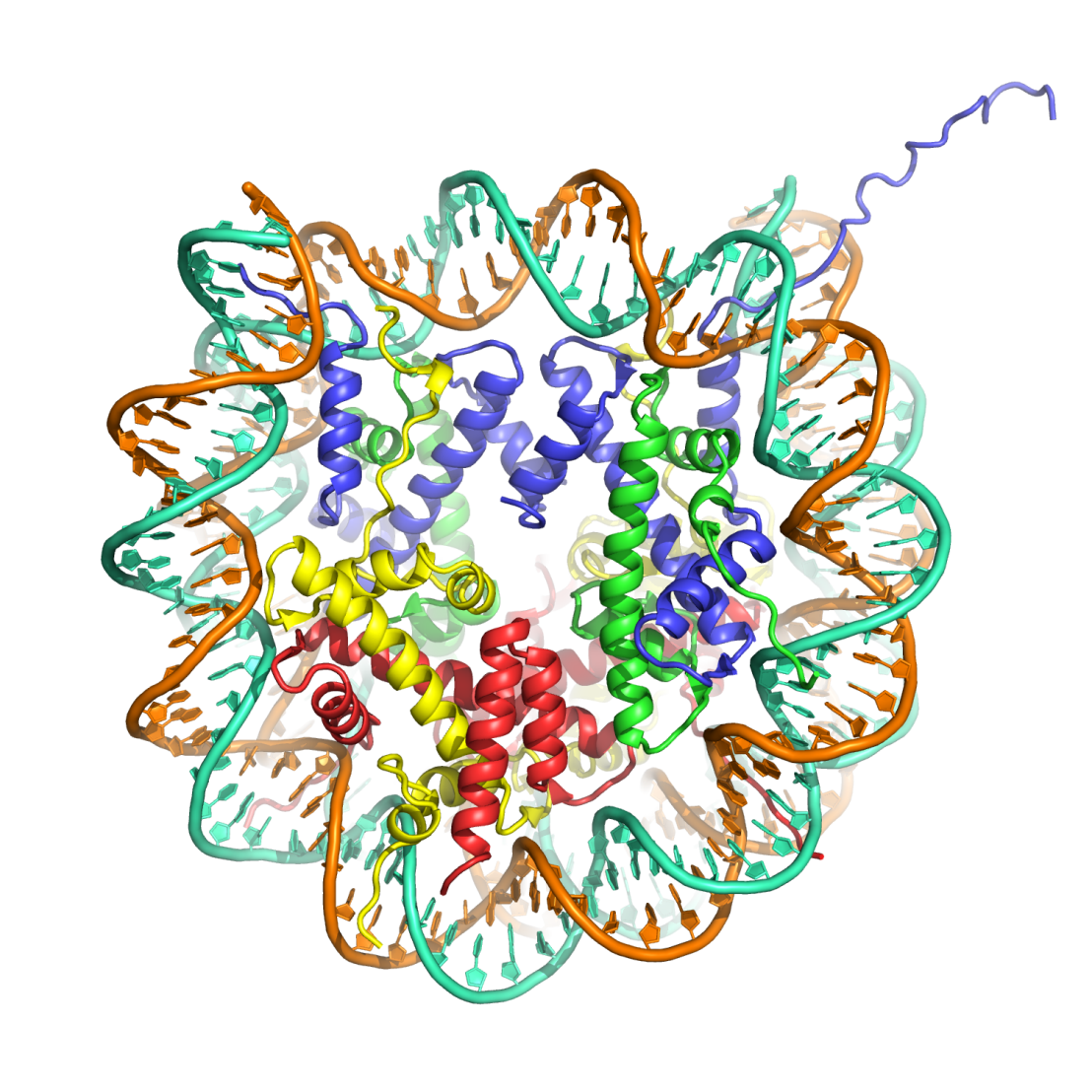
We study the molecular structure of chromatin and its associated factors by X-ray crystallography, cryoelectron microscopy and biochemical analysis. The regulation of gene expression through chromatin remodeling and modification are of particular interest. Read more about our research.
Group Wider

Based on new structure determinations of biologically interesting macromolecules we improve existing and develop new NMR experiments. Furthermore, for proteins with known X-ray structure we develop new assignment methods that also work for large molecules where conventional techniques fail. We are interested in aqueous as well as in nonaqueous solutions. In the latter enzymes cannot only be active but also exhibit intriguing new properties, however, the phenomenon is not yet understood in detail since there are very limited structural and dynamic data at atomic resolution. Read more about our research.